Population density affects sexual selection in an insect model
Part 2: Testing the effect of density on mating behaviour and reproductive success
Lennart Winkler1, Ronja
Eilhardt1 & Tim Janicke1,2
1Applied Zoology, Technical University Dresden
2Centre d’Écologie Fonctionnelle et Évolutive, UMR 5175,
CNRS, Université de Montpellier
Last updated: 2023-05-04
Checks: 7 0
Knit directory:
Density_and_sexual_selection_2023/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210613) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version fe2f665. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/index3.Rmd) and HTML
(docs/index3.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | fe2f665 | LennartWinkler | 2023-05-04 | wflow_publish(all = T) |
| html | af34366 | LennartWinkler | 2023-04-27 | Build site. |
| Rmd | e4a0f09 | LennartWinkler | 2023-04-27 | wflow_publish(all = T) |
| html | b81114e | LennartWinkler | 2023-04-20 | Build site. |
| Rmd | fe8c52c | LennartWinkler | 2023-04-20 | wflow_publish(all = T) |
| html | fe8c52c | LennartWinkler | 2023-04-20 | wflow_publish(all = T) |
| html | 55df7ff | LennartWinkler | 2023-04-12 | Build site. |
| html | b0a3f15 | LennartWinkler | 2023-04-12 | Build site. |
| Rmd | c8bd540 | Lennart Winkler | 2023-04-12 | set up |
| html | c8bd540 | Lennart Winkler | 2023-04-12 | set up |
| html | 39e340c | LennartWinkler | 2022-08-13 | new |
| Rmd | f54f022 | LennartWinkler | 2022-08-13 | wflow_publish(republish = TRUE, all = T) |
| html | f54f022 | LennartWinkler | 2022-08-13 | wflow_publish(republish = TRUE, all = T) |
| Rmd | f89f7c1 | LennartWinkler | 2022-08-10 | Build site. |
| html | f89f7c1 | LennartWinkler | 2022-08-10 | Build site. |
Supplementary material reporting R code for the manuscript ‘Population density affects sexual selection in an insect model’.
Part 2: Testing the effect of density on mating behaviour and reproductive success
Here we tested the effect that the treatments (group and arena size)
had on the mating behaviour and reproductive success of focal
beetles.
Behavioural variables:
- Copulation attempts
-
Copulations per encounter
- Number of partners
- Number
copulations
- Copulations per partner
- Copulation duration
Load and prepare data
Before we started the analyses, we loaded all necessary packages and data.
rm(list = ls()) # Clear work environment
# Load R-packages ####
list_of_packages=cbind('ggeffects','ggplot2','gridExtra','lme4','lmerTest','readr','dplyr','EnvStats','cowplot','gridGraphics','car','RColorBrewer','boot','data.table','base','ICC','knitr')
lapply(list_of_packages, require, character.only = TRUE)
# Load data set ####
D_data=read_delim("./data/Data_Winkler_et_al_2023_Denstiy.csv",";", escape_double = FALSE, trim_ws = TRUE)
# Set factors and levels for factors
D_data$Week=as.factor(D_data$Week)
D_data$Sex=as.factor(D_data$Sex)
D_data$Gr_size=as.factor(D_data$Gr_size)
D_data$Gr_size <- factor(D_data$Gr_size, levels=c("SG","LG"))
D_data$Arena=as.factor(D_data$Arena)
## Subset data set ####
### Data according to denstiy ####
D_data_0.26=D_data[D_data$Treatment=='D = 0.26',]
D_data_0.52=D_data[D_data$Treatment=='D = 0.52',]
D_data_0.67=D_data[D_data$Treatment=='D = 0.67',]
D_data_1.33=D_data[D_data$Treatment=='D = 1.33',]
### Subset data by sex ####
D_data_m=D_data[D_data$Sex=='M',]
D_data_f=D_data[D_data$Sex=='F',]
### Calculate data relativized within treatment and sex ####
# Small group + large Area
D_data_0.26=D_data[D_data$Treatment=='D = 0.26',]
D_data_0.26$rel_m_RS=NA
D_data_0.26$rel_m_prop_RS=NA
D_data_0.26$rel_m_cMS=NA
D_data_0.26$rel_m_InSuc=NA
D_data_0.26$rel_m_feSuc=NA
D_data_0.26$rel_m_pFec=NA
D_data_0.26$rel_m_PS=NA
D_data_0.26$rel_m_pFec_compl=NA
D_data_0.26$rel_f_RS=NA
D_data_0.26$rel_f_prop_RS=NA
D_data_0.26$rel_f_cMS=NA
D_data_0.26$rel_f_fec_pMate=NA
D_data_0.26$rel_m_RS=D_data_0.26$m_RS/mean(D_data_0.26$m_RS,na.rm=T)
D_data_0.26$rel_m_prop_RS=D_data_0.26$m_prop_RS/mean(D_data_0.26$m_prop_RS,na.rm=T)
D_data_0.26$rel_m_cMS=D_data_0.26$m_cMS/mean(D_data_0.26$m_cMS,na.rm=T)
D_data_0.26$rel_m_InSuc=D_data_0.26$m_InSuc/mean(D_data_0.26$m_InSuc,na.rm=T)
D_data_0.26$rel_m_feSuc=D_data_0.26$m_feSuc/mean(D_data_0.26$m_feSuc,na.rm=T)
D_data_0.26$rel_m_pFec=D_data_0.26$m_pFec/mean(D_data_0.26$m_pFec,na.rm=T)
D_data_0.26$rel_m_PS=D_data_0.26$m_PS/mean(D_data_0.26$m_PS,na.rm=T)
D_data_0.26$rel_m_pFec_compl=D_data_0.26$m_pFec_compl/mean(D_data_0.26$m_pFec_compl,na.rm=T)
D_data_0.26$rel_f_RS=D_data_0.26$f_RS/mean(D_data_0.26$f_RS,na.rm=T)
D_data_0.26$rel_f_prop_RS=D_data_0.26$f_prop_RS/mean(D_data_0.26$f_prop_RS,na.rm=T)
D_data_0.26$rel_f_cMS=D_data_0.26$f_cMS/mean(D_data_0.26$f_cMS,na.rm=T)
D_data_0.26$rel_f_fec_pMate=D_data_0.26$f_fec_pMate/mean(D_data_0.26$f_fec_pMate,na.rm=T)
# Large group + large Area
D_data_0.52=D_data[D_data$Treatment=='D = 0.52',]
#Relativize data
D_data_0.52$rel_m_RS=NA
D_data_0.52$rel_m_prop_RS=NA
D_data_0.52$rel_m_cMS=NA
D_data_0.52$rel_m_InSuc=NA
D_data_0.52$rel_m_feSuc=NA
D_data_0.52$rel_m_pFec=NA
D_data_0.52$rel_m_PS=NA
D_data_0.52$rel_m_pFec_compl=NA
D_data_0.52$rel_f_RS=NA
D_data_0.52$rel_f_prop_RS=NA
D_data_0.52$rel_f_cMS=NA
D_data_0.52$rel_f_fec_pMate=NA
D_data_0.52$rel_m_RS=D_data_0.52$m_RS/mean(D_data_0.52$m_RS,na.rm=T)
D_data_0.52$rel_m_prop_RS=D_data_0.52$m_prop_RS/mean(D_data_0.52$m_prop_RS,na.rm=T)
D_data_0.52$rel_m_cMS=D_data_0.52$m_cMS/mean(D_data_0.52$m_cMS,na.rm=T)
D_data_0.52$rel_m_InSuc=D_data_0.52$m_InSuc/mean(D_data_0.52$m_InSuc,na.rm=T)
D_data_0.52$rel_m_feSuc=D_data_0.52$m_feSuc/mean(D_data_0.52$m_feSuc,na.rm=T)
D_data_0.52$rel_m_pFec=D_data_0.52$m_pFec/mean(D_data_0.52$m_pFec,na.rm=T)
D_data_0.52$rel_m_PS=D_data_0.52$m_PS/mean(D_data_0.52$m_PS,na.rm=T)
D_data_0.52$rel_m_pFec_compl=D_data_0.52$m_pFec_compl/mean(D_data_0.52$m_pFec_compl,na.rm=T)
D_data_0.52$rel_f_RS=D_data_0.52$f_RS/mean(D_data_0.52$f_RS,na.rm=T)
D_data_0.52$rel_f_prop_RS=D_data_0.52$f_prop_RS/mean(D_data_0.52$f_prop_RS,na.rm=T)
D_data_0.52$rel_f_cMS=D_data_0.52$f_cMS/mean(D_data_0.52$f_cMS,na.rm=T)
D_data_0.52$rel_f_fec_pMate=D_data_0.52$f_fec_pMate/mean(D_data_0.52$f_fec_pMate,na.rm=T)
# Small group + small Area
D_data_0.67=D_data[D_data$Treatment=='D = 0.67',]
#Relativize data
D_data_0.67$rel_m_RS=NA
D_data_0.67$rel_m_prop_RS=NA
D_data_0.67$rel_m_cMS=NA
D_data_0.67$rel_m_InSuc=NA
D_data_0.67$rel_m_feSuc=NA
D_data_0.67$rel_m_pFec=NA
D_data_0.67$rel_m_PS=NA
D_data_0.67$rel_m_pFec_compl=NA
D_data_0.67$rel_f_RS=NA
D_data_0.67$rel_f_prop_RS=NA
D_data_0.67$rel_f_cMS=NA
D_data_0.67$rel_f_fec_pMate=NA
D_data_0.67$rel_m_RS=D_data_0.67$m_RS/mean(D_data_0.67$m_RS,na.rm=T)
D_data_0.67$rel_m_prop_RS=D_data_0.67$m_prop_RS/mean(D_data_0.67$m_prop_RS,na.rm=T)
D_data_0.67$rel_m_cMS=D_data_0.67$m_cMS/mean(D_data_0.67$m_cMS,na.rm=T)
D_data_0.67$rel_m_InSuc=D_data_0.67$m_InSuc/mean(D_data_0.67$m_InSuc,na.rm=T)
D_data_0.67$rel_m_feSuc=D_data_0.67$m_feSuc/mean(D_data_0.67$m_feSuc,na.rm=T)
D_data_0.67$rel_m_pFec=D_data_0.67$m_pFec/mean(D_data_0.67$m_pFec,na.rm=T)
D_data_0.67$rel_m_PS=D_data_0.67$m_PS/mean(D_data_0.67$m_PS,na.rm=T)
D_data_0.67$rel_m_pFec_compl=D_data_0.67$m_pFec_compl/mean(D_data_0.67$m_pFec_compl,na.rm=T)
D_data_0.67$rel_f_RS=D_data_0.67$f_RS/mean(D_data_0.67$f_RS,na.rm=T)
D_data_0.67$rel_f_prop_RS=D_data_0.67$f_prop_RS/mean(D_data_0.67$f_prop_RS,na.rm=T)
D_data_0.67$rel_f_cMS=D_data_0.67$f_cMS/mean(D_data_0.67$f_cMS,na.rm=T)
D_data_0.67$rel_f_fec_pMate=D_data_0.67$f_fec_pMate/mean(D_data_0.67$f_fec_pMate,na.rm=T)
# Large group + small Area
D_data_1.33=D_data[D_data$Treatment=='D = 1.33',]
#Relativize data
D_data_1.33$rel_m_RS=NA
D_data_1.33$rel_m_prop_RS=NA
D_data_1.33$rel_m_cMS=NA
D_data_1.33$rel_m_InSuc=NA
D_data_1.33$rel_m_feSuc=NA
D_data_1.33$rel_m_pFec=NA
D_data_1.33$rel_m_PS=NA
D_data_1.33$rel_m_pFec_compl=NA
D_data_1.33$rel_f_RS=NA
D_data_1.33$rel_f_prop_RS=NA
D_data_1.33$rel_f_cMS=NA
D_data_1.33$rel_f_fec_pMate=NA
D_data_1.33$rel_m_RS=D_data_1.33$m_RS/mean(D_data_1.33$m_RS,na.rm=T)
D_data_1.33$rel_m_prop_RS=D_data_1.33$m_prop_RS/mean(D_data_1.33$m_prop_RS,na.rm=T)
D_data_1.33$rel_m_cMS=D_data_1.33$m_cMS/mean(D_data_1.33$m_cMS,na.rm=T)
D_data_1.33$rel_m_InSuc=D_data_1.33$m_InSuc/mean(D_data_1.33$m_InSuc,na.rm=T)
D_data_1.33$rel_m_feSuc=D_data_1.33$m_feSuc/mean(D_data_1.33$m_feSuc,na.rm=T)
D_data_1.33$rel_m_pFec=D_data_1.33$m_pFec/mean(D_data_1.33$m_pFec,na.rm=T)
D_data_1.33$rel_m_PS=D_data_1.33$m_PS/mean(D_data_1.33$m_PS,na.rm=T)
D_data_1.33$rel_m_pFec_compl=D_data_1.33$m_pFec_compl/mean(D_data_1.33$m_pFec_compl,na.rm=T)
D_data_1.33$rel_f_RS=D_data_1.33$f_RS/mean(D_data_1.33$f_RS,na.rm=T)
D_data_1.33$rel_f_prop_RS=D_data_1.33$f_prop_RS/mean(D_data_1.33$f_prop_RS,na.rm=T)
D_data_1.33$rel_f_cMS=D_data_1.33$f_cMS/mean(D_data_1.33$f_cMS,na.rm=T)
D_data_1.33$rel_f_fec_pMate=D_data_1.33$f_fec_pMate/mean(D_data_1.33$f_fec_pMate,na.rm=T)
### Reduce treatments to arena and population size ####
# Arena size
D_data_Large_arena=rbind(D_data_0.26,D_data_0.52)
D_data_Small_arena=rbind(D_data_0.67,D_data_1.33)
# Population size
D_data_Small_pop=rbind(D_data_0.26,D_data_0.67)
D_data_Large_pop=rbind(D_data_0.52,D_data_1.33)
## Set figure schemes ####
# Set color-sets for figures
colpal=brewer.pal(4, 'Dark2')
colpal2=c("#b2182b","#2166AC")
colpal3=brewer.pal(4, 'Paired')
# Set theme for ggplot2 figures
fig_theme=theme(panel.border = element_blank(),
plot.margin = margin(0,2.2,0,0.2,"cm"),
plot.title = element_text(hjust = 0.5),
panel.background = element_blank(),
legend.key=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
legend.position = c(1.25, 0.8),
plot.tag.position=c(0.01,0.98),
legend.title = element_blank(),
legend.text = element_text(colour="black", size=10),
axis.line.x = element_line(colour = "black", size = 1),
axis.line.y = element_line(colour = "black", size = 1),
axis.text.x = element_text(face="plain", color="black", size=16, angle=0),
axis.text.y = element_text(face="plain", color="black", size=16, angle=0),
axis.title.x = element_text(size=16,face="plain", margin = margin(r=0,10,0,0)),
axis.title.y = element_text(size=16,face="plain", margin = margin(r=10,0,0,0)),
axis.ticks = element_line(size = 1),
axis.ticks.length = unit(.3, "cm"))
## Create customized functions for analysis ####
# Create function to calculate standard error and upper/lower standard deviation
standard_error <- function(x) sd(x,na.rm=T) / sqrt(length(na.exclude(x)))
upper_CI <- function(x) mean(x,na.rm=T)+((standard_error(x))*qnorm(0.975))
lower_CI <- function(x) mean(x,na.rm=T)-((standard_error(x))*qnorm(0.975))
upper_SD <- function(x) mean(x,na.rm=T)+(sd(x)/2)
lower_SD <- function(x) mean(x,na.rm=T)-(sd(x)/2)Model the effect of density on mating behaviour
Copulation attempts
Males
First, we calculated means and SE for each treatment: Mean number of copulation attempts in small groups (SE) = 11.04 (0.97)
mean(D_data$m_Total_Encounters[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_Total_Encounters[D_data$Gr_size=='SG'])Mean number of copulation attempts in large groups (SE) = 7.64 (0.49)
mean(D_data$m_Total_Encounters[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_Total_Encounters[D_data$Gr_size=='LG'])Mean number of copulation attempts in large arena size (SE) = 8.98 (0.72)
mean(D_data$m_Total_Encounters[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_Total_Encounters[D_data$Arena=='Large'])Mean number of copulation attempts in small arena size (SE) = 9.33 (0.8)
mean(D_data$m_Total_Encounters[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_Total_Encounters[D_data$Arena=='Small'])GLM for copulation attempts including interaction between group and arena size treatment:
mod3=glm(m_Total_Encounters~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod3,type=2)Analysis of Deviance Table (Type II tests)
Response: m_Total_Encounters
LR Chisq Df Pr(>Chisq)
Gr_size 11.4368 1 0.0007201 ***
Arena 0.0520 1 0.8195638
Gr_size:Arena 0.2226 1 0.6370758
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for copulation attempts excluding interaction between group and arena size treatment:
mod3.1=glm(m_Total_Encounters~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod3.1)
Call:
glm(formula = m_Total_Encounters ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.8812 -1.4114 -0.2269 0.8185 5.1613
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.41598 0.09612 25.135 < 2e-16 ***
Gr_sizeLG -0.37295 0.10996 -3.392 0.000992 ***
ArenaSmall -0.02515 0.10978 -0.229 0.819291
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 2.780109)
Null deviance: 302.24 on 103 degrees of freedom
Residual deviance: 269.83 on 101 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod3.1,type=2)Analysis of Deviance Table (Type II tests)
Response: m_Total_Encounters
LR Chisq Df Pr(>Chisq)
Gr_size 11.5351 1 0.0006829 ***
Arena 0.0525 1 0.8188029
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Females
We calculated means and SE for each treatment: Mean number of copulation attempts in small groups (SE) = 8.8 (0.68)
mean(D_data$f_Total_Encounters[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_Total_Encounters[D_data$Gr_size=='SG'])Mean number of copulation attempts in large groups (SE) = 6.36 (0.44)
mean(D_data$f_Total_Encounters[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_Total_Encounters[D_data$Gr_size=='LG'])Mean number of copulation attempts in large arena size (SE) = 8.1 (0.65)
mean(D_data$f_Total_Encounters[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_Total_Encounters[D_data$Arena=='Large'])Mean number of copulation attempts in small arena size (SE) = 7.25 (0.58)
mean(D_data$f_Total_Encounters[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_Total_Encounters[D_data$Arena=='Small'])GLM for copulation attempts including interaction between group and arena size treatment:
mod4=glm(f_Total_Encounters~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod4,type=2)Analysis of Deviance Table (Type II tests)
Response: f_Total_Encounters
LR Chisq Df Pr(>Chisq)
Gr_size 8.0855 1 0.004462 **
Arena 0.2888 1 0.590966
Gr_size:Arena 0.5385 1 0.463060
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for copulation attempts excluding interaction between group and arena size treatment:
mod4.1=glm(f_Total_Encounters~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod4.1)
Call:
glm(formula = f_Total_Encounters ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-4.2437 -1.0822 -0.2262 0.9494 3.6489
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.19774 0.07997 27.483 < 2e-16 ***
Gr_sizeLG -0.31505 0.11178 -2.819 0.00586 **
ArenaSmall -0.05846 0.10867 -0.538 0.59183
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 2.169405)
Null deviance: 241.05 on 98 degrees of freedom
Residual deviance: 221.13 on 96 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod4.1,type=2)Analysis of Deviance Table (Type II tests)
Response: f_Total_Encounters
LR Chisq Df Pr(>Chisq)
Gr_size 8.1140 1 0.004392 **
Arena 0.2899 1 0.590313
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Copulations per attempt
Males
We calculated means and SE for each treatment: Number of copulations per attempt in small groups (SE) = 0.32 (0.03)
mean(D_data$m_TotMatings[D_data$Gr_size=='SG']/(D_data$m_TotMatings[D_data$Gr_size=='SG']+D_data$m_Attempts_number[D_data$Gr_size=='SG']),na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Gr_size=='SG']/(D_data$m_TotMatings[D_data$Gr_size=='SG']+D_data$m_Attempts_number[D_data$Gr_size=='SG']))Number of copulations per attempt in large groups (SE) = 0.29 (0.02)
mean(D_data$m_TotMatings[D_data$Gr_size=='LG']/(D_data$m_TotMatings[D_data$Gr_size=='LG']+D_data$m_Attempts_number[D_data$Gr_size=='LG']),na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Gr_size=='LG']/(D_data$m_TotMatings[D_data$Gr_size=='LG']+D_data$m_Attempts_number[D_data$Gr_size=='LG']))Number of copulations per attempt in large arena size (SE) = 0.31 (0.03)
mean(D_data$m_TotMatings[D_data$Arena=='Large']/(D_data$m_TotMatings[D_data$Arena=='Large']+D_data$m_Attempts_number[D_data$Arena=='Large']),na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Arena=='Large']/(D_data$m_TotMatings[D_data$Arena=='Large']+D_data$m_Attempts_number[D_data$Arena=='Large']))Number of copulations per attempt in small arena size (SE) = 0.3 (0.03)
mean(D_data$m_TotMatings[D_data$Arena=='Small']/(D_data$m_TotMatings[D_data$Arena=='Small']+D_data$m_Attempts_number[D_data$Arena=='Small']),na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Arena=='Small']/(D_data$m_TotMatings[D_data$Arena=='Small']+D_data$m_Attempts_number[D_data$Arena=='Small']))GLM for copulations per attempt including interaction between group and arena size treatment:
mod5=glm(cbind(m_TotMatings,m_Attempts_number)~Gr_size*Arena,data=D_data,family = quasibinomial)
Anova(mod5,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(m_TotMatings, m_Attempts_number)
LR Chisq Df Pr(>Chisq)
Gr_size 0.18848 1 0.6642
Arena 0.28809 1 0.5914
Gr_size:Arena 0.11436 1 0.7352GLM for copulations per attempt excluding interaction between group and arena size treatment:
mod5.1=glm(cbind(m_TotMatings,m_Attempts_number)~Gr_size+Arena,data=D_data,family = quasibinomial)
summary(mod5.1)
Call:
glm(formula = cbind(m_TotMatings, m_Attempts_number) ~ Gr_size +
Arena, family = quasibinomial, data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.6291 -0.8482 -0.0073 0.7556 4.4019
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.94176 0.16198 -5.814 7.17e-08 ***
Gr_sizeLG 0.08134 0.18650 0.436 0.664
ArenaSmall -0.10058 0.18667 -0.539 0.591
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 1.623469)
Null deviance: 180.01 on 103 degrees of freedom
Residual deviance: 179.10 on 101 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 4Anova(mod5.1,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(m_TotMatings, m_Attempts_number)
LR Chisq Df Pr(>Chisq)
Gr_size 0.19011 1 0.6628
Arena 0.29059 1 0.5898Females
We calculated means and SE for each treatment: Number of copulations per attempt in small groups (SE) = 0.37 (0.04)
mean(D_data$f_TotMatings[D_data$Gr_size=='SG']/(D_data$f_TotMatings[D_data$Gr_size=='SG']+D_data$f_Attempts_number[D_data$Gr_size=='SG']),na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Gr_size=='SG']/(D_data$f_TotMatings[D_data$Gr_size=='SG']+D_data$f_Attempts_number[D_data$Gr_size=='SG']))Number of copulations per attempt in large groups (SE) = 0.32 (0.04)
mean(D_data$f_TotMatings[D_data$Gr_size=='LG']/(D_data$f_TotMatings[D_data$Gr_size=='LG']+D_data$f_Attempts_number[D_data$Gr_size=='LG']),na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Gr_size=='LG']/(D_data$f_TotMatings[D_data$Gr_size=='LG']+D_data$f_Attempts_number[D_data$Gr_size=='LG']))Number of copulations per attempt in large arena size (SE) = 0.39 (0.04)
mean(D_data$f_TotMatings[D_data$Arena=='Large']/(D_data$f_TotMatings[D_data$Arena=='Large']+D_data$f_Attempts_number[D_data$Arena=='Large']),na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Arena=='Large']/(D_data$f_TotMatings[D_data$Arena=='Large']+D_data$f_Attempts_number[D_data$Arena=='Large']))Number of copulations per attempt in small arena size (SE) = 0.3 (0.04)
mean(D_data$f_TotMatings[D_data$Arena=='Small']/(D_data$f_TotMatings[D_data$Arena=='Small']+D_data$f_Attempts_number[D_data$Arena=='Small']),na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Arena=='Small']/(D_data$f_TotMatings[D_data$Arena=='Small']+D_data$f_Attempts_number[D_data$Arena=='Small']))GLM for copulations per attempt including interaction between group and arena size treatment:
mod6=glm(cbind(f_TotMatings,f_Attempts_number)~Gr_size*Arena,data=D_data,family = quasibinomial)
Anova(mod6,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(f_TotMatings, f_Attempts_number)
LR Chisq Df Pr(>Chisq)
Gr_size 0.00646 1 0.9359
Arena 1.62223 1 0.2028
Gr_size:Arena 1.63103 1 0.2016GLM for copulations per attempt excluding interaction between group and arena size treatment:
mod6.1=glm(cbind(f_TotMatings,f_Attempts_number)~Gr_size+Arena,data=D_data,family = quasibinomial)
summary(mod6.1)
Call:
glm(formula = cbind(f_TotMatings, f_Attempts_number) ~ Gr_size +
Arena, family = quasibinomial, data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.88927 -0.92593 -0.04092 0.98199 2.72954
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.64091 0.14996 -4.274 4.57e-05 ***
Gr_sizeLG -0.01687 0.21061 -0.080 0.936
ArenaSmall -0.25990 0.20536 -1.266 0.209
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 1.67619)
Null deviance: 183.58 on 97 degrees of freedom
Residual deviance: 180.78 on 95 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 4Anova(mod6.1,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(f_TotMatings, f_Attempts_number)
LR Chisq Df Pr(>Chisq)
Gr_size 0.00642 1 0.9362
Arena 1.61064 1 0.2044Number of partners
Males
We calculated means and SE for each treatment: Mean number of partners in small groups (SE) = 1.63 (0.11)
mean(D_data$m_cMS[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_cMS[D_data$Gr_size=='SG'])Mean number of partners in large groups (SE) = 1.69 (0.14)
mean(D_data$m_cMS[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_cMS[D_data$Gr_size=='LG'])Mean number of partners in large arena size (SE) = 1.69 (0.14)
mean(D_data$m_cMS[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_cMS[D_data$Arena=='Large'])Mean number of partners in small arena size (SE) = 1.63 (0.12)
mean(D_data$m_cMS[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_cMS[D_data$Arena=='Small'])GLM for number of partners including interaction between group and arena size treatment
mod7=glm(m_cMS~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod7,type=2)Analysis of Deviance Table (Type II tests)
Response: m_cMS
LR Chisq Df Pr(>Chisq)
Gr_size 0.071366 1 0.7894
Arena 0.069055 1 0.7927
Gr_size:Arena 0.004674 1 0.9455GLM for number of partners excluding interaction between group and arena size treatment
mod7.1=glm(m_cMS~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod7.1)
Call:
glm(formula = m_cMS ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.8491 -0.5529 0.2162 0.2670 2.0373
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.50567 0.10574 4.782 5.92e-06 ***
Gr_sizeLG 0.03063 0.11416 0.268 0.789
ArenaSmall -0.02995 0.11346 -0.264 0.792
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 0.5381483)
Null deviance: 65.695 on 103 degrees of freedom
Residual deviance: 65.603 on 101 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod7.1,type=2)Analysis of Deviance Table (Type II tests)
Response: m_cMS
LR Chisq Df Pr(>Chisq)
Gr_size 0.072077 1 0.7883
Arena 0.069742 1 0.7917Number of partners
Females
We calculated means and SE for each treatment: Mean number of partners in small groups (SE) = 1.69 (0.12)
mean(D_data$f_cMS[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_cMS[D_data$Gr_size=='SG'])Mean number of partners in large groups (SE) = 1.73 (0.2)
mean(D_data$f_cMS[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_cMS[D_data$Gr_size=='LG'])Mean number of partners in large arena size (SE) = 1.86 (0.15)
mean(D_data$f_cMS[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_cMS[D_data$Arena=='Large'])Mean number of partners in small arena size (SE) = 1.54 (0.17)
mean(D_data$f_cMS[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_cMS[D_data$Arena=='Small'])GLM for number of partners including interaction between group and arena size treatment:
mod8=glm(f_cMS~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod8,type=2)Analysis of Deviance Table (Type II tests)
Response: f_cMS
LR Chisq Df Pr(>Chisq)
Gr_size 0.21523 1 0.6427
Arena 2.22688 1 0.1356
Gr_size:Arena 1.06086 1 0.3030GLM for number of partners excluding interaction between group and arena size treatment:
mod8.1=glm(f_cMS~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod8.1)
Call:
glm(formula = f_cMS ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.9676 -0.6646 0.1318 0.3968 2.1580
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.59849 0.10194 5.871 6.19e-08 ***
Gr_sizeLG 0.06204 0.13393 0.463 0.644
ArenaSmall -0.19970 0.13457 -1.484 0.141
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 0.7318014)
Null deviance: 89.305 on 98 degrees of freedom
Residual deviance: 87.650 on 96 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod8.1,type=2)Analysis of Deviance Table (Type II tests)
Response: f_cMS
LR Chisq Df Pr(>Chisq)
Gr_size 0.2142 1 0.6435
Arena 2.2162 1 0.1366Number copulations
Males
We calculated means and SE for each treatment: Mean number of copulations in small groups (SE) = 2.98 (0.31)
mean(D_data$m_TotMatings[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Gr_size=='SG'])Mean number of copulations in large groups (SE) = 2.21 (0.21)
mean(D_data$m_TotMatings[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Gr_size=='LG'])Mean number of copulations in large arena size (SE) = 2.6 (0.29)
mean(D_data$m_TotMatings[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Arena=='Large'])Mean number of copulations in small arena size (SE) = 2.49 (0.22)
mean(D_data$m_TotMatings[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_TotMatings[D_data$Arena=='Small'])GLM for number of copulations including interaction between group and arena size treatment:
mod9=glm(m_TotMatings~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod9,type=2)Analysis of Deviance Table (Type II tests)
Response: m_TotMatings
LR Chisq Df Pr(>Chisq)
Gr_size 4.8747 1 0.02725 *
Arena 0.4538 1 0.50056
Gr_size:Arena 0.4756 1 0.49043
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for number of copulations excluding interaction between group and arena size treatment:
mod9.1=glm(m_TotMatings~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod9.1)
Call:
glm(formula = m_TotMatings ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.5067 -0.9610 -0.1962 0.4978 3.0725
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.14475 0.12532 9.135 7.21e-15 ***
Gr_sizeLG -0.31599 0.14338 -2.204 0.0298 *
ArenaSmall -0.09653 0.14375 -0.672 0.5034
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 1.321896)
Null deviance: 146.28 on 103 degrees of freedom
Residual deviance: 139.74 on 101 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod9.1,type=2)Analysis of Deviance Table (Type II tests)
Response: m_TotMatings
LR Chisq Df Pr(>Chisq)
Gr_size 4.8548 1 0.02757 *
Arena 0.4519 1 0.50143
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Females
We calculated means and SE for each treatment: Mean number of copulations in small groups (SE) = 2.83 (0.3)
mean(D_data$f_TotMatings[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Gr_size=='SG'])Mean number of copulations in large groups (SE) = 1.98 (0.25)
mean(D_data$f_TotMatings[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Gr_size=='LG'])Mean number of copulations in large arena size (SE) = 2.78 (0.29)
mean(D_data$f_TotMatings[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Arena=='Large'])Mean number of copulations in small arena size (SE) = 2.08 (0.27)
mean(D_data$f_TotMatings[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_TotMatings[D_data$Arena=='Small'])GLM for number of copulations including interaction between group and arena size treatment:
mod10=glm(f_TotMatings~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod10,type=2)Analysis of Deviance Table (Type II tests)
Response: f_TotMatings
LR Chisq Df Pr(>Chisq)
Gr_size 3.6584 1 0.05579 .
Arena 2.0796 1 0.14928
Gr_size:Arena 0.3714 1 0.54226
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for number of copulations excluding interaction between group and arena size treatment:
mod10.1=glm(f_TotMatings~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod10.1)
Call:
glm(formula = f_TotMatings ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.4900 -1.0500 -0.1716 0.6617 3.6136
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.1314 0.1161 9.742 5.33e-16 ***
Gr_sizeLG -0.3193 0.1683 -1.898 0.0607 .
ArenaSmall -0.2372 0.1648 -1.440 0.1532
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 1.550111)
Null deviance: 171.30 on 98 degrees of freedom
Residual deviance: 160.59 on 96 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod10.1,type=2)Analysis of Deviance Table (Type II tests)
Response: f_TotMatings
LR Chisq Df Pr(>Chisq)
Gr_size 3.6862 1 0.05486 .
Arena 2.0954 1 0.14774
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Copulations per partner
Males
We calculated means and SE for each treatment: Copulations per partner in small groups (SE) = 1.79 (0.12)
mean(D_data$m_meanCop[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_meanCop[D_data$Gr_size=='SG'])Copulations per partner in large groups (SE) = 1.33 (0.09)
mean(D_data$m_meanCop[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_meanCop[D_data$Gr_size=='LG'])Copulations per partner in large arena size (SE) = 1.51 (0.11)
mean(D_data$m_meanCop[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_meanCop[D_data$Arena=='Large'])Copulations per partner in small arena size (SE) = 1.57 (0.1)
mean(D_data$m_meanCop[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_meanCop[D_data$Arena=='Small'])GLM for copulations per partner including interaction between group and arena size treatment:
mod11=glm(cbind(m_cMS,m_TotMatings)~Gr_size*Arena,data=D_data,family = quasibinomial)
Anova(mod11,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(m_cMS, m_TotMatings)
LR Chisq Df Pr(>Chisq)
Gr_size 17.5682 1 2.772e-05 ***
Arena 0.4728 1 0.4917
Gr_size:Arena 1.6703 1 0.1962
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for copulations per partner excluding interaction between group and arena size treatment:
mod11.1=glm(cbind(m_cMS,m_TotMatings)~Gr_size+Arena,data=D_data,family = quasibinomial)
summary(mod11.1)
Call:
glm(formula = cbind(m_cMS, m_TotMatings) ~ Gr_size + Arena, family = quasibinomial,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.1268 -0.1276 0.1645 0.2889 0.7831
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.63273 0.07417 -8.531 3.02e-13 ***
Gr_sizeLG 0.34336 0.08231 4.172 6.90e-05 ***
ArenaSmall 0.05651 0.08236 0.686 0.494
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 0.1717382)
Null deviance: 18.871 on 93 degrees of freedom
Residual deviance: 15.865 on 91 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 3Anova(mod11.1,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(m_cMS, m_TotMatings)
LR Chisq Df Pr(>Chisq)
Gr_size 17.4924 1 2.885e-05 ***
Arena 0.4708 1 0.4926
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Females
We calculated means and SE for each treatment: Copulations per partner in small groups (SE) = 1.61 (0.1)
mean(D_data$f_meanCop[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_meanCop[D_data$Gr_size=='SG'])Copulations per partner in large groups (SE) = 1.13 (0.06)
mean(D_data$f_meanCop[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_meanCop[D_data$Gr_size=='LG'])Copulations per partner in large arena size (SE) = 1.42 (0.07)
mean(D_data$f_meanCop[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_meanCop[D_data$Arena=='Large'])Copulations per partner in small arena size (SE) = 1.4 (0.13)
mean(D_data$f_meanCop[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_meanCop[D_data$Arena=='Small'])GLM for copulations per partner including interaction between group and arena size treatment:
mod12=glm(cbind(f_cMS,f_TotMatings)~Gr_size*Arena,data=D_data,family = quasibinomial)
Anova(mod12,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(f_cMS, f_TotMatings)
LR Chisq Df Pr(>Chisq)
Gr_size 25.4678 1 4.498e-07 ***
Arena 0.0412 1 0.8391
Gr_size:Arena 0.2179 1 0.6407
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for copulations per partner excluding interaction between group and arena size treatment:
mod12.1=glm(cbind(f_cMS,f_TotMatings)~Gr_size+Arena,data=D_data,family = quasibinomial)
summary(mod12.1)
Call:
glm(formula = cbind(f_cMS, f_TotMatings) ~ Gr_size + Arena, family = quasibinomial,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.11605 -0.14258 0.09933 0.15315 1.23097
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.52471 0.05415 -9.690 3.45e-15 ***
Gr_sizeLG 0.38418 0.07577 5.070 2.46e-06 ***
ArenaSmall 0.01544 0.07563 0.204 0.839
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 0.1311458)
Null deviance: 14.763 on 83 degrees of freedom
Residual deviance: 11.140 on 81 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 3Anova(mod12.1,type=2)Analysis of Deviance Table (Type II tests)
Response: cbind(f_cMS, f_TotMatings)
LR Chisq Df Pr(>Chisq)
Gr_size 25.7447 1 3.897e-07 ***
Arena 0.0417 1 0.8383
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Copulation duration
Males
We calculated means and SE for each treatment: Copulation duration in small groups (SE) = 78.98 (4.88)
mean(D_data$m_MatingDuration_av[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_MatingDuration_av[D_data$Gr_size=='SG'])Copulation duration in large groups (SE) = 74.8 (4.7)
mean(D_data$m_MatingDuration_av[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_MatingDuration_av[D_data$Gr_size=='LG'])Copulation duration in large arena size (SE) = 77.41 (5.15)
mean(D_data$m_MatingDuration_av[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_MatingDuration_av[D_data$Arena=='Large'])Copulation duration in small arena size (SE) = 75.95 (4.35)
mean(D_data$m_MatingDuration_av[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_MatingDuration_av[D_data$Arena=='Small'])GLM for copulation duration including interaction between group and arena size treatment:
mod13=glm(m_MatingDuration_av~Gr_size*Arena,data=D_data,family = gaussian)
Anova(mod13,type=2)Analysis of Deviance Table (Type II tests)
Response: m_MatingDuration_av
LR Chisq Df Pr(>Chisq)
Gr_size 0.43450 1 0.5098
Arena 0.11108 1 0.7389
Gr_size:Arena 0.04691 1 0.8285GLM for copulation duration excluding interaction between group and arena size treatment:
mod13.1=glm(m_MatingDuration_av~Gr_size+Arena,data=D_data,family = gaussian)
summary(mod13.1)
Call:
glm(formula = m_MatingDuration_av ~ Gr_size + Arena, family = gaussian,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-44.333 -21.092 -9.408 10.981 111.783
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 80.333 6.457 12.441 <2e-16 ***
Gr_sizeLG -4.615 6.965 -0.663 0.509
ArenaSmall -2.327 6.946 -0.335 0.738
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 1091.532)
Null deviance: 99859 on 93 degrees of freedom
Residual deviance: 99329 on 91 degrees of freedom
(109 Beobachtungen als fehlend gelöscht)
AIC: 929.27
Number of Fisher Scoring iterations: 2Anova(mod13.1,type=2)Analysis of Deviance Table (Type II tests)
Response: m_MatingDuration_av
LR Chisq Df Pr(>Chisq)
Gr_size 0.43909 1 0.5076
Arena 0.11225 1 0.7376Females
We calculated means and SE for each treatment: Copulation duration in small groups (SE) = 84.13 (6.9)
mean(D_data$f_MatingDuration_av[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_MatingDuration_av[D_data$Gr_size=='SG'])Copulation duration in large groups (SE) = 68.47 (3.35)
mean(D_data$f_MatingDuration_av[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_MatingDuration_av[D_data$Gr_size=='LG'])Copulation duration in large arena size (SE) = 80.27 (7.07)
mean(D_data$f_MatingDuration_av[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_MatingDuration_av[D_data$Arena=='Large'])Copulation duration in small arena size (SE) = 74.55 (4.38)
mean(D_data$f_MatingDuration_av[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_MatingDuration_av[D_data$Arena=='Small'])GLM for copulation duration including interaction between group and arena size treatment:
mod14=glm(f_MatingDuration_av~Gr_size*Arena,data=D_data,family = gaussian)
Anova(mod14,type=2)Analysis of Deviance Table (Type II tests)
Response: f_MatingDuration_av
LR Chisq Df Pr(>Chisq)
Gr_size 2.90863 1 0.08811 .
Arena 0.17022 1 0.67991
Gr_size:Arena 1.20855 1 0.27162
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1GLM for copulation duration excluding interaction between group and arena size treatment:
mod14.1=glm(f_MatingDuration_av~Gr_size+Arena,data=D_data,family = gaussian)
summary(mod14.1)
Call:
glm(formula = f_MatingDuration_av ~ Gr_size + Arena, family = gaussian,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-45.913 -21.965 -7.868 11.425 252.462
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 85.538 6.587 12.986 <2e-16 ***
Gr_sizeLG -15.145 8.892 -1.703 0.0924 .
ArenaSmall -3.625 8.797 -0.412 0.6814
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 1555.818)
Null deviance: 129651 on 82 degrees of freedom
Residual deviance: 124465 on 80 degrees of freedom
(120 Beobachtungen als fehlend gelöscht)
AIC: 850.52
Number of Fisher Scoring iterations: 2Anova(mod14.1,type=2)Analysis of Deviance Table (Type II tests)
Response: f_MatingDuration_av
LR Chisq Df Pr(>Chisq)
Gr_size 2.90107 1 0.08852 .
Arena 0.16978 1 0.68031
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Reproductive success
Males
We calculated means and SE for each treatment: Reproductive success in small groups (SE) = 64.2 (9.45)
mean(D_data$m_RS[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$m_RS[D_data$Gr_size=='SG'])Reproductive success in large groups (SE) = 49.29 (6.14)
mean(D_data$m_RS[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$m_RS[D_data$Gr_size=='LG'])Reproductive success in large arena size (SE) = 51.24 (6.2)
mean(D_data$m_RS[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$m_RS[D_data$Arena=='Large'])Reproductive success in small arena size (SE) = 61.1 (9.19)
mean(D_data$m_RS[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$m_RS[D_data$Arena=='Small'])GLM for reproductive success including interaction between group and arena size treatment:
mod15=glm(m_RS~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod15,type=2)Analysis of Deviance Table (Type II tests)
Response: m_RS
LR Chisq Df Pr(>Chisq)
Gr_size 1.56524 1 0.2109
Arena 0.49398 1 0.4822
Gr_size:Arena 1.22773 1 0.2678GLM for reproductive success excluding interaction between group and arena size treatment:
mod15.1=glm(m_RS~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod15.1)
Call:
glm(formula = m_RS ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-11.6566 -9.6552 -0.6194 4.4727 21.1646
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.0831 0.1767 23.111 <2e-16 ***
Gr_sizeLG -0.2413 0.1932 -1.249 0.215
ArenaSmall 0.1355 0.1933 0.701 0.485
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 52.69429)
Null deviance: 6253.5 on 103 degrees of freedom
Residual deviance: 6126.4 on 101 degrees of freedom
(99 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod15.1,type=2)Analysis of Deviance Table (Type II tests)
Response: m_RS
LR Chisq Df Pr(>Chisq)
Gr_size 1.55751 1 0.2120
Arena 0.49155 1 0.4832Females
We calculated means and SE for each treatment: Reproductive success in small groups (SE) = 51.59 (6.91)
mean(D_data$f_RS[D_data$Gr_size=='SG'],na.rm=T)
standard_error(D_data$f_RS[D_data$Gr_size=='SG'])Reproductive success in large groups (SE) = 48.93 (7.11)
mean(D_data$f_RS[D_data$Gr_size=='LG'],na.rm=T)
standard_error(D_data$f_RS[D_data$Gr_size=='LG'])Reproductive success in large arena size (SE) = 52.76 (7.04)
mean(D_data$f_RS[D_data$Arena=='Large'],na.rm=T)
standard_error(D_data$f_RS[D_data$Arena=='Large'])Reproductive success in small arena size (SE) = 47.85 (7)
mean(D_data$f_RS[D_data$Arena=='Small'],na.rm=T)
standard_error(D_data$f_RS[D_data$Arena=='Small'])GLM for reproductive success including interaction between group and arena size treatment:
mod16=glm(f_RS~Gr_size*Arena,data=D_data,family = quasipoisson)
Anova(mod16,type=2)Analysis of Deviance Table (Type II tests)
Response: f_RS
LR Chisq Df Pr(>Chisq)
Gr_size 0.03445 1 0.8528
Arena 0.20764 1 0.6486
Gr_size:Arena 1.54223 1 0.2143GLM for reproductive success excluding interaction between group and arena size treatment:
mod16.1=glm(f_RS~Gr_size+Arena,data=D_data,family = quasipoisson)
summary(mod16.1)
Call:
glm(formula = f_RS ~ Gr_size + Arena, family = quasipoisson,
data = D_data)
Deviance Residuals:
Min 1Q Median 3Q Max
-10.3437 -9.8817 0.8721 5.6151 9.5213
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.97960 0.15361 25.907 <2e-16 ***
Gr_sizeLG -0.03737 0.20234 -0.185 0.854
ArenaSmall -0.09137 0.20157 -0.453 0.651
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 48.91479)
Null deviance: 6268.7 on 98 degrees of freedom
Residual deviance: 6255.2 on 96 degrees of freedom
(104 Beobachtungen als fehlend gelöscht)
AIC: NA
Number of Fisher Scoring iterations: 5Anova(mod16.1,type=2)Analysis of Deviance Table (Type II tests)
Response: f_RS
LR Chisq Df Pr(>Chisq)
Gr_size 0.03416 1 0.8534
Arena 0.20588 1 0.6500Calculate adjusted p-values
We calculated adjusted p-values within sex and treatment using the false discovery rate correction (FDR). Adjusted p-values for population size treatment effect in females:
tab1=data.table(round(p.adjust(cbind(Anova(mod4.1,type=2)[1,3],Anova(mod6.1,type=2)[1,3],Anova(mod8.1,type=2)[1,3],Anova(mod10.1,type=2)[1,3],Anova(mod12.1,type=2)[1,3],Anova(mod14.1,type=2)[1,3],Anova(mod16.1,type=2)[1,3]), method = 'fdr'),digit=3)) # Compute FDR corrected p-values
traits=rbind('Copulation attempts','Copulations per attempt','Number of partners','Number copulations','Copulations per partner','Copulation duration','Reproductive success')
tab1= cbind(traits,tab1)
colnames(tab1)= cbind('Trait','Adj. p-value')
tab1 Trait Adj. p-value
1: Copulation attempts 0.015
2: Copulations per attempt 0.936
3: Number of partners 0.901
4: Number copulations 0.128
5: Copulations per partner 0.000
6: Copulation duration 0.155
7: Reproductive success 0.936Adjusted p-values for arena size treatment effect in females:
tab2=data.table(round(p.adjust(cbind(Anova(mod4.1,type=2)[2,3],Anova(mod6.1,type=2)[2,3],Anova(mod8.1,type=2)[2,3],Anova(mod10.1,type=2)[2,3],Anova(mod12.1,type=2)[2,3],Anova(mod14.1,type=2)[2,3],Anova(mod16.1,type=2)[2,3]), method = 'fdr'),digit=3)) # Compute FDR corrected p-values
tab2= cbind(traits,tab2)
colnames(tab2)= cbind('Trait','Adj. p-value')
tab2 Trait Adj. p-value
1: Copulation attempts 0.794
2: Copulations per attempt 0.477
3: Number of partners 0.477
4: Number copulations 0.477
5: Copulations per partner 0.838
6: Copulation duration 0.794
7: Reproductive success 0.794Adjusted p-values for population size treatment effect in males:
tab3=data.table(round(p.adjust(cbind(Anova(mod3.1,type=2)[1,3],Anova(mod5.1,type=2)[1,3],Anova(mod7.1,type=2)[1,3],Anova(mod9.1,type=2)[1,3],Anova(mod11.1,type=2)[1,3],Anova(mod13.1,type=2)[1,3],Anova(mod15.1,type=2)[1,3]), method = 'fdr'),digit=3)) # Compute FDR corrected p-values
tab3= cbind(traits,tab3)
colnames(tab3)= cbind('Trait','Adj. p-value')
tab3 Trait Adj. p-value
1: Copulation attempts 0.002
2: Copulations per attempt 0.773
3: Number of partners 0.788
4: Number copulations 0.064
5: Copulations per partner 0.000
6: Copulation duration 0.711
7: Reproductive success 0.371Adjusted p-values for population size treatment effect in males:
tab4=data.table(round(p.adjust(cbind(Anova(mod3.1,type=2)[2,3],Anova(mod5.1,type=2)[2,3],Anova(mod7.1,type=2)[2,3],Anova(mod9.1,type=2)[2,3],Anova(mod11.1,type=2)[2,3],Anova(mod13.1,type=2)[2,3],Anova(mod15.1,type=2)[2,3]), method = 'fdr'),digit=3)) # Compute FDR corrected p-values
tab4= cbind(traits,tab4)
colnames(tab4)= cbind('Trait','Adj. p-value')
tab4 Trait Adj. p-value
1: Copulation attempts 0.819
2: Copulations per attempt 0.819
3: Number of partners 0.819
4: Number copulations 0.819
5: Copulations per partner 0.819
6: Copulation duration 0.819
7: Reproductive success 0.819Figures for mating behaviour and reproductive success (Figure S2-S4)
Here we plotted the mating behaviour and reproductive success per treatment and sex.
## Figures for mating behaviour and reproductive success (Figure S2-S4) ####
# Create factor for treatment categories
D_data$TreatCgroup <- factor(paste(D_data$Sex,D_data$Gr_size, sep=" "), levels = c("F SG", "F LG", "M SG",'M LG'))
D_data$TreatCarena <- factor(paste(D_data$Sex,D_data$Arena, sep=" "), levels = c("F Large", "F Small", "M Large",'M Small'))
p3<-ggplot(D_data, aes(x=Sex, y=as.numeric(Total_Encounters),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('Sex')+ylab("Copulation attempts")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,30)+labs(tag = "C")+
annotate("text",label='n =',x=0.55,y=30,size=4)+
annotate("text",label='54',x=0.78,y=30,size=4)+
annotate("text",label='45',x=1.23,y=30,size=4)+
annotate("text",label='46',x=1.78,y=30,size=4)+
annotate("text",label='58',x=2.23,y=30,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p3.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(Total_Encounters),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('Sex')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,30)+labs(tag = "D")+
annotate("text",label='n =',x=0.55,y=30,size=4)+
annotate("text",label='51',x=0.78,y=30,size=4)+
annotate("text",label='48',x=1.23,y=30,size=4)+
annotate("text",label='55',x=1.78,y=30,size=4)+
annotate("text",label='49',x=2.23,y=30,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
### Plot: Copulations per attempt ####
p4<-ggplot(D_data, aes(x=Sex, y=as.numeric(Prop_MS),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('')+ylab("Copulations per attempt")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,1.1)+labs(tag = "A")+
annotate("text",label='n =',x=0.55,y=1.1,size=4)+
annotate("text",label='54',x=0.78,y=1.1,size=4)+
annotate("text",label='45',x=1.23,y=1.1,size=4)+
annotate("text",label='46',x=1.78,y=1.1,size=4)+
annotate("text",label='58',x=2.23,y=1.1,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p4.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(Prop_MS),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,1.1)+labs(tag = "B")+
annotate("text",label='n =',x=0.55,y=1.1,size=4)+
annotate("text",label='51',x=0.78,y=1.1,size=4)+
annotate("text",label='48',x=1.23,y=1.1,size=4)+
annotate("text",label='55',x=1.78,y=1.1,size=4)+
annotate("text",label='49',x=2.23,y=1.1,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
### Plot: Mating partners focal ####
p5<-ggplot(D_data, aes(x=Sex, y=as.numeric(MatingPartners_number),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('')+ylab("Number of partners")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,5.4)+labs(tag = "C")+
annotate("text",label='n =',x=0.55,y=5.4,size=4)+
annotate("text",label='54',x=0.78,y=5.4,size=4)+
annotate("text",label='45',x=1.23,y=5.4,size=4)+
annotate("text",label='46',x=1.78,y=5.4,size=4)+
annotate("text",label='58',x=2.23,y=5.4,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p5.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(MatingPartners_number),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,5.4)+labs(tag = "D")+
annotate("text",label='n =',x=0.55,y=5.4,size=4)+
annotate("text",label='51',x=0.78,y=5.4,size=4)+
annotate("text",label='48',x=1.23,y=5.4,size=4)+
annotate("text",label='55',x=1.78,y=5.4,size=4)+
annotate("text",label='49',x=2.23,y=5.4,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
### Plot: Number copulations ####
p6<-ggplot(D_data, aes(x=Sex, y=as.numeric(Matings_number),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('')+ylab("Number of copulations")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,12)+labs(tag = "A")+
annotate("text",label='n =',x=0.55,y=12,size=4)+
annotate("text",label='54',x=0.78,y=12,size=4)+
annotate("text",label='45',x=1.23,y=12,size=4)+
annotate("text",label='46',x=1.78,y=12,size=4)+
annotate("text",label='58',x=2.23,y=12,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p6.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(Matings_number),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+labs(tag = "B")+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,12)+
annotate("text",label='n =',x=0.55,y=12,size=4)+
annotate("text",label='51',x=0.78,y=12,size=4)+
annotate("text",label='48',x=1.23,y=12,size=4)+
annotate("text",label='55',x=1.78,y=12,size=4)+
annotate("text",label='49',x=2.23,y=12,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
###Plot: Copulations per partner ####
p7<-ggplot(D_data, aes(x=Sex, y=as.numeric(meanCop),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('Sex')+ylab("Copulations per partner")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ labs(tag = "C")+ylim(1,4.5)+
annotate("text",label='n =',x=0.55,y=4.5,size=4)+
annotate("text",label='54',x=0.78,y=4.5,size=4)+
annotate("text",label='45',x=1.23,y=4.5,size=4)+
annotate("text",label='46',x=1.78,y=4.5,size=4)+
annotate("text",label='58',x=2.23,y=4.5,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p7.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(meanCop),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('Sex')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(1,4.5)+labs(tag = "D")+
annotate("text",label='n =',x=0.55,y=4.5,size=4)+
annotate("text",label='51',x=0.78,y=4.5,size=4)+
annotate("text",label='48',x=1.23,y=4.5,size=4)+
annotate("text",label='55',x=1.78,y=4.5,size=4)+
annotate("text",label='49',x=2.23,y=4.5,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
### Plot: Copulation duration ####
p8<-ggplot(D_data, aes(x=Sex, y=as.numeric(MatingDuration_av),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('Sex')+ylab("Mean copulation duration")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,390)+labs(tag = "E")+
annotate("text",label='n =',x=0.55,y=390,size=4)+
annotate("text",label='49',x=0.78,y=390,size=4)+
annotate("text",label='34',x=1.23,y=390,size=4)+
annotate("text",label='43',x=1.78,y=390,size=4)+
annotate("text",label='51',x=2.23,y=390,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p8.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(MatingDuration_av),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('Sex')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,390)+labs(tag = "F")+
annotate("text",label='n =',x=0.55,y=390,size=4)+
annotate("text",label='46',x=0.78,y=390,size=4)+
annotate("text",label='37',x=1.23,y=390,size=4)+
annotate("text",label='49',x=1.78,y=390,size=4)+
annotate("text",label='45',x=2.23,y=390,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
###Plot: Reproductive success ####
p9<-ggplot(D_data, aes(x=Sex, y=as.numeric(Total_N_MTP1),fill=TreatCgroup, col=TreatCgroup,alpha=TreatCgroup)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small group','Large group','Small group','Large group'))+
xlab('Sex')+ylab("Number of offspring")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,320)+labs(tag = "A")+
annotate("text",label='n =',x=0.55,y=320,size=4)+
annotate("text",label='54',x=0.78,y=320,size=4)+
annotate("text",label='45',x=1.23,y=320,size=4)+
annotate("text",label='46',x=1.78,y=320,size=4)+
annotate("text",label='58',x=2.23,y=320,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_theme
p9.2<-ggplot(D_data, aes(x=Sex, y=as.numeric(Total_N_MTP1),fill=TreatCarena, col=TreatCarena,alpha=TreatCarena)) +
geom_point(position=position_jitterdodge(jitter.width=0.5,jitter.height = 0,dodge.width=1.2),shape=19, size = 2)+
stat_summary(fun.min =lower_CI ,
fun.max = upper_CI ,fun = mean,
position=position_dodge2(0.3),col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, linewidth = 1.15)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("white","white","white","white"),alpha=c(1,1,1,1),show.legend = F, stroke = 0,linewidth = 1.2)+
stat_summary(fun = mean,
position=position_dodge2(0.3), size = 1,col=c("#b2182b","#b2182b","#2166AC","#2166AC"),alpha=c(0.5,0.75,0.5,0.75),show.legend = F, stroke = 0,linewidth = 1.2)+
scale_color_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_fill_manual(values=c(colpal2[1],colpal2[1],colpal2[2],colpal2[2]),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
scale_alpha_manual(values=c(0.5,0.75,0.5,0.75),name = "Treatment", labels = c('Small arena','Large arena','Small arena','Large arena'))+
xlab('Sex')+ylab("")+ggtitle('')+ theme(plot.title = element_text(hjust = 0.5))+
scale_x_discrete(labels = c('Female','Male'),drop=FALSE)+ ylim(0,320)+labs(tag = "B")+
annotate("text",label='n =',x=0.55,y=320,size=4)+
annotate("text",label='51',x=0.78,y=320,size=4)+
annotate("text",label='48',x=1.23,y=320,size=4)+
annotate("text",label='55',x=1.78,y=320,size=4)+
annotate("text",label='49',x=2.23,y=320,size=4)+
guides(colour = guide_legend(override.aes = list(size=4)))+
fig_themeFigure S2
#Behaviour 1 (Figure S2)
Figure_S2<-grid.arrange(p3+labs(tag = "A")+xlab('')+theme(legend.position = c(1.25, 0.8),plot.margin = unit(c(0.2,3.5,0,0.1), "cm"),legend.text = element_text(size=9)),p3.2+
labs(tag = "B")+xlab('')+theme(legend.position = c(1.25, 0.8),plot.margin = unit(c(0.2,3.5,0,0.1), "cm"),legend.text = element_text(size=9)),
p4+labs(tag = "C")+xlab('')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),p4.2+labs(tag = "D")+xlab('')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),
p5+labs(tag = "E")+xlab('Sex')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),p5.2+labs(tag = "F")+xlab('Sex')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm"))
,nrow = 3,ncol=2)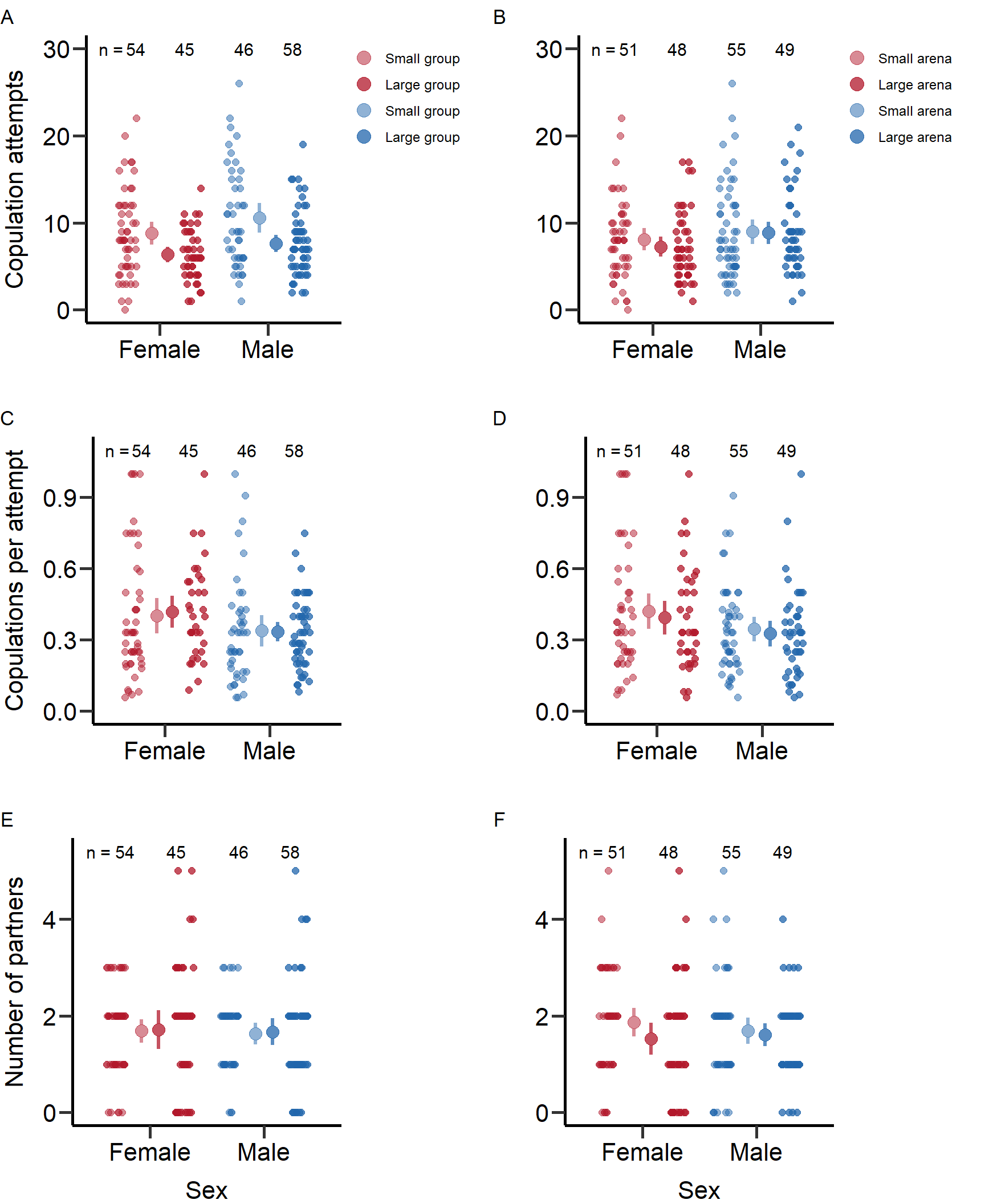
Figure S2: Mating behavior of males and females under low and high density manipulation via group (left) and arena size (right). Bars indicate means and 95% CI.
Figure_S2 = plot_grid(Figure_S2, ncol=1, rel_heights=c(0.1, 1)) # rel_heights values control title marginsFigure S3
#Behavior 2 (Figure S3)
Figure_S3<-grid.arrange(p6+labs(tag = "A")+xlab('')+theme(legend.position = c(1.25, 0.8),plot.margin = unit(c(0.2,3.5,0,0.1), "cm"),legend.text = element_text(size=9)),p6.2+labs(tag = "B")+xlab('')+theme(legend.position = c(1.25, 0.8),plot.margin = unit(c(0.2,3.5,0,0.1), "cm"),legend.text = element_text(size=9)),
p7+labs(tag = "C")+xlab('')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),p7.2+labs(tag = "D")+xlab('')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),
p8+labs(tag = "E")+xlab('Sex')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),p8.2+labs(tag = "F")+xlab('Sex')+theme(legend.position = 'none',plot.margin = unit(c(0.2,3.5,0,0.1), "cm")),nrow = 3,ncol=2)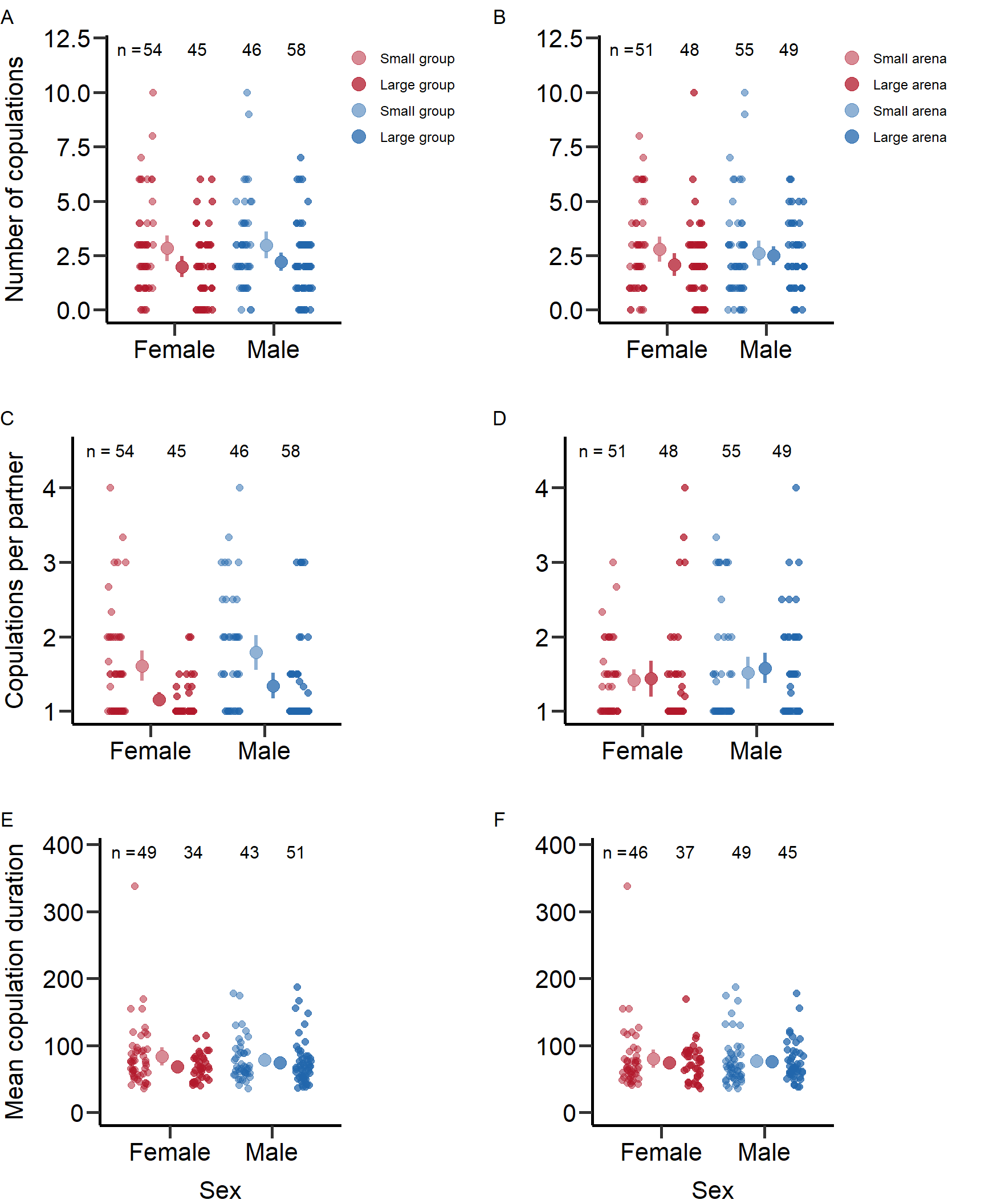
Figure S3: Mating behavior of males and females under low and high density manipulation via group (left) and arena size (right). Bars indicate means and 95% CI.
Figure_S3 = plot_grid(Figure_S3, ncol=1, rel_heights=c(0.1, 1)) # rel_heights values control title marginsFigure S4
# Reproductive success (Figure S4)
Figure_S4<-grid.arrange(grobs = list(p9+theme(legend.position = c(1.2, 0.8),plot.margin = unit(c(0.2,4,0,0.3), "cm")),p9.2+theme(legend.position = c(1.2, 0.8),plot.margin = unit(c(0.2,4,0,0.3), "cm"))), nrow = 1,ncol=2, widths=c(2.3, 2.3))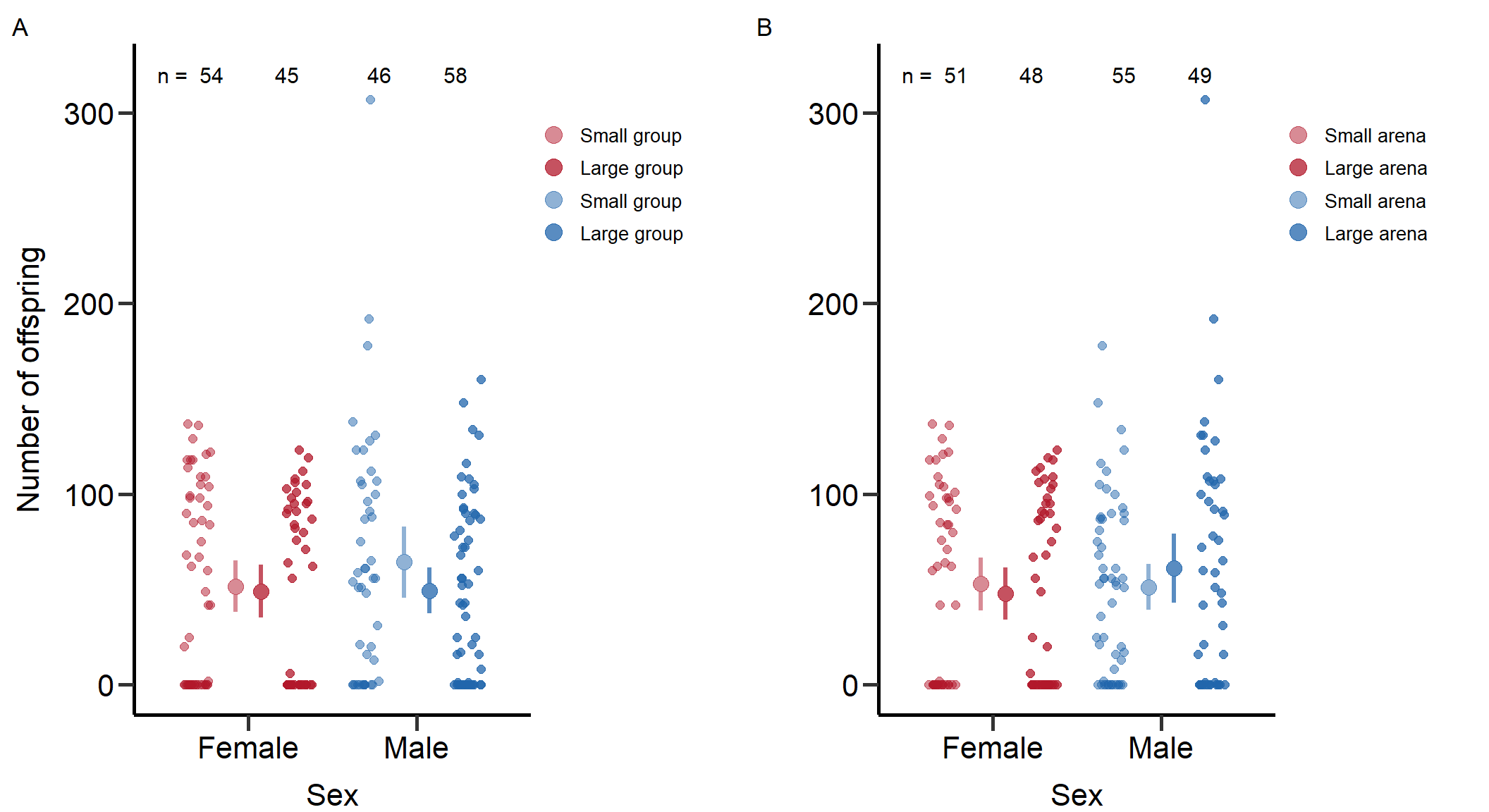
Figure S4: Reproductive success of males and females under density manipulation via group (left) and arena size (right). Bars indicate means and 95% CI.
Figure_S4 = plot_grid(Figure_S4, ncol=1, rel_heights=c(0.1, 1)) # rel_heights values control title margins
sessionInfo()R version 4.2.0 (2022-04-22 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=German_Germany.utf8 LC_CTYPE=German_Germany.utf8
[3] LC_MONETARY=German_Germany.utf8 LC_NUMERIC=C
[5] LC_TIME=German_Germany.utf8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] knitr_1.42 ICC_2.4.0 data.table_1.14.8 boot_1.3-28
[5] RColorBrewer_1.1-3 car_3.1-1 carData_3.0-5 gridGraphics_0.5-1
[9] cowplot_1.1.1 EnvStats_2.7.0 dplyr_1.1.0 readr_2.1.4
[13] lmerTest_3.1-3 lme4_1.1-31 Matrix_1.5-3 gridExtra_2.3
[17] ggplot2_3.4.1 ggeffects_1.2.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.5 sass_0.4.5 bit64_4.0.5
[4] vroom_1.6.1 jsonlite_1.8.4 splines_4.2.0
[7] bslib_0.4.2 getPass_0.2-2 highr_0.10
[10] yaml_2.3.7 numDeriv_2016.8-1.1 pillar_1.8.1
[13] lattice_0.20-45 glue_1.6.2 digest_0.6.31
[16] promises_1.2.0.1 minqa_1.2.5 colorspace_2.1-0
[19] htmltools_0.5.4 httpuv_1.6.9 pkgconfig_2.0.3
[22] scales_1.2.1 processx_3.8.0 whisker_0.4.1
[25] later_1.3.0 tzdb_0.3.0 git2r_0.31.0
[28] tibble_3.2.0 generics_0.1.3 farver_2.1.1
[31] ellipsis_0.3.2 cachem_1.0.7 withr_2.5.0
[34] cli_3.6.1 magrittr_2.0.3 crayon_1.5.2
[37] evaluate_0.20 ps_1.7.2 fs_1.6.1
[40] fansi_1.0.4 nlme_3.1-157 MASS_7.3-56
[43] tools_4.2.0 hms_1.1.2 lifecycle_1.0.3
[46] stringr_1.5.0 munsell_0.5.0 callr_3.7.3
[49] compiler_4.2.0 jquerylib_0.1.4 rlang_1.0.6
[52] nloptr_2.0.3 rstudioapi_0.14 labeling_0.4.2
[55] rmarkdown_2.20 gtable_0.3.1 abind_1.4-5
[58] R6_2.5.1 fastmap_1.1.1 bit_4.0.5
[61] utf8_1.2.3 rprojroot_2.0.3 stringi_1.7.12
[64] parallel_4.2.0 Rcpp_1.0.10 vctrs_0.5.2
[67] tidyselect_1.2.0 xfun_0.37